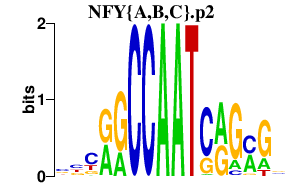
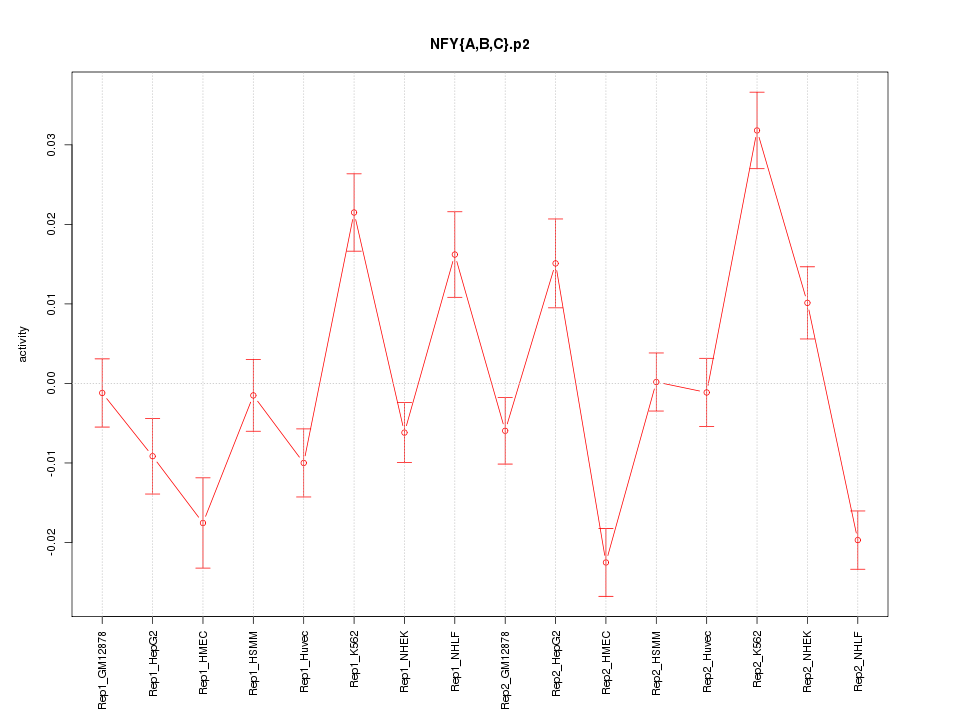
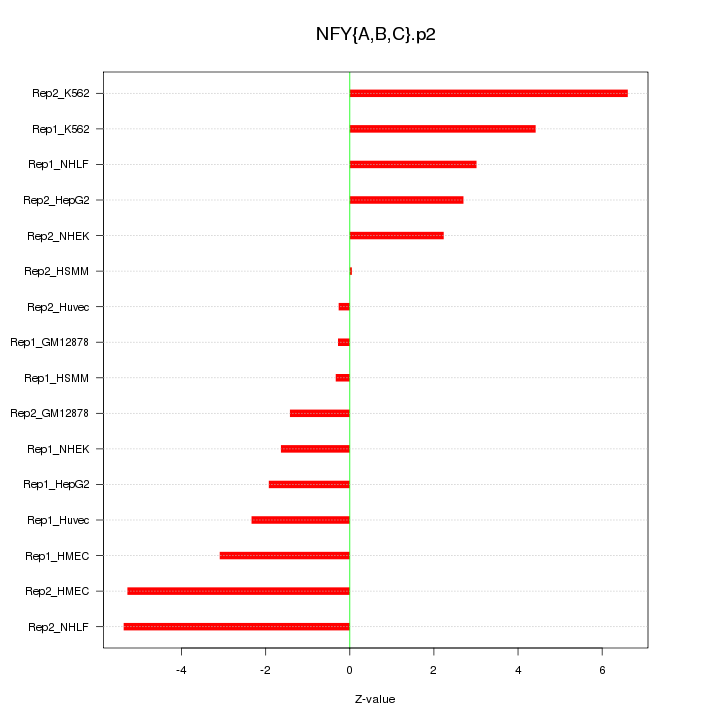
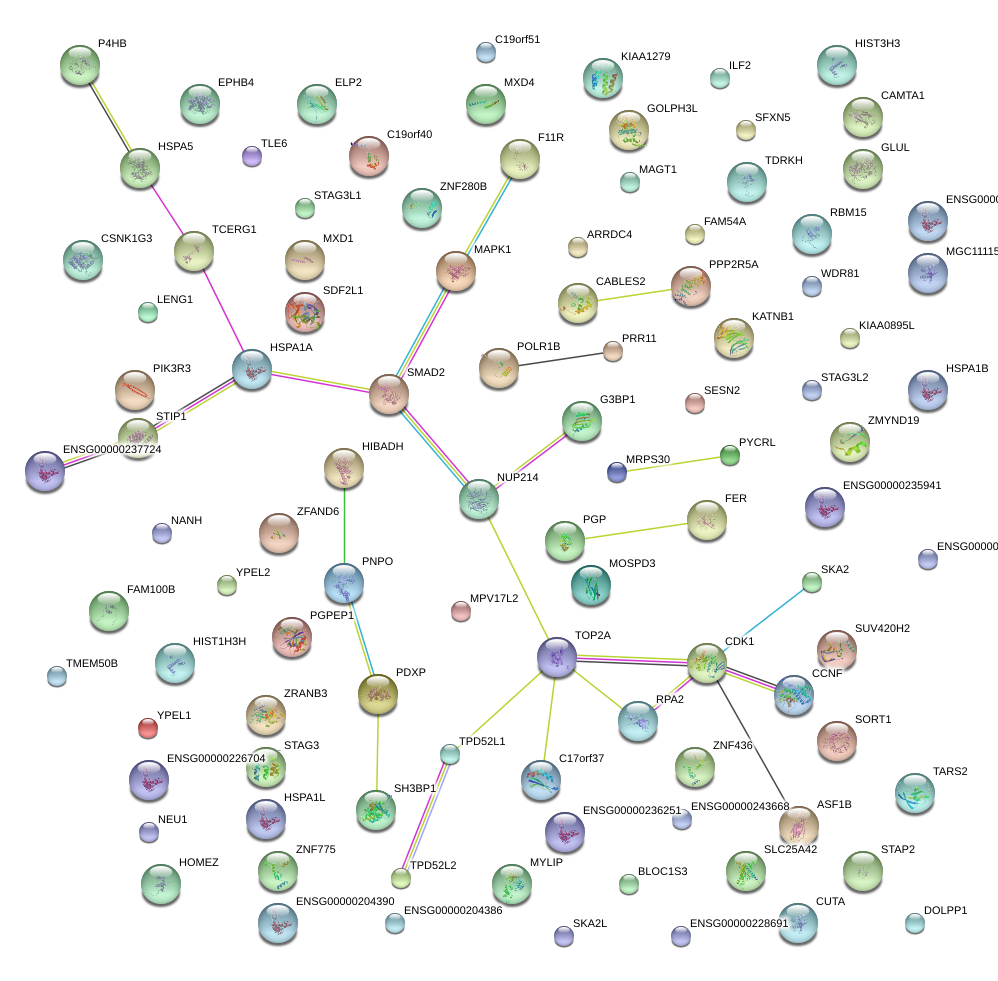
|
chr7_-_27669029
|
3.289
|
NM_152740
|
HIBADH
|
3-hydroxyisobutyrate dehydrogenase
|
|
chr19_-_4293718
|
3.247
|
|
STAP2
|
signal transducing adaptor family member 2
|
|
chr22_-_20419975
|
3.137
|
NM_013313
|
YPEL1
|
yippee-like 1 (Drosophila)
|
|
chr8_-_144762765
|
3.016
|
NM_023078
|
PYCRL
|
pyrroline-5-carboxylate reductase-like
|
|
chr17_-_35827568
|
2.847
|
NM_001067
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa
|
|
chr2_+_69995656
|
2.491
|
NM_002357
|
MXD1
|
MAX dimerization protein 1
|
|
chr6_-_31938572
|
2.469
|
NM_000434
|
NEU1
|
sialidase 1 (lysosomal sialidase)
|
|
chr1_+_110683467
|
2.456
|
NM_022768
|
RBM15
|
RNA binding motif protein 15
|
|
chr17_+_54587882
|
2.411
|
|
PRR11
|
proline rich 11
|
|
chr18_-_43711452
|
2.383
|
NM_001003652
NM_001135937
|
SMAD2
|
SMAD family member 2
|
|
chr1_+_210525397
|
2.371
|
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha
|
|
chr1_-_151910057
|
2.354
|
|
ILF2
|
interleukin enhancer binding factor 2, 45kDa
|
|
chr1_-_151910052
|
2.354
|
|
ILF2
|
interleukin enhancer binding factor 2, 45kDa
|
|
chr2_+_69995740
|
2.331
|
|
MXD1
|
MAX dimerization protein 1
|
|
chr1_-_151910080
|
2.330
|
|
ILF2
|
interleukin enhancer binding factor 2, 45kDa
|
|
chr1_-_151910041
|
2.307
|
|
ILF2
|
interleukin enhancer binding factor 2, 45kDa
|
|
chr19_-_59355246
|
2.299
|
NM_024316
|
LENG1
|
leukocyte receptor cluster (LRC) member 1
|
|
chr6_+_31903374
|
2.193
|
NM_005346
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chrX_-_77037563
|
2.181
|
NM_032121
|
MAGT1
|
magnesium transporter 1
|
|
chr17_-_54587581
|
2.151
|
NM_001100595
NM_182620
|
SKA2
|
spindle and kinetochore associated complex subunit 2
|
|
chr19_-_60369731
|
2.131
|
NM_178837
|
C19orf51
|
chromosome 19 open reading frame 51
|
|
chr22_+_20326541
|
2.114
|
NM_022044
|
SDF2L1
|
stromal cell-derived factor 2-like 1
|
|
chr22_+_20326572
|
2.055
|
|
SDF2L1
|
stromal cell-derived factor 2-like 1
|
|
chr5_+_145807059
|
2.049
|
NM_001040006
NM_006706
|
TCERG1
|
transcription elongation regulator 1
|
|
chr19_+_60543032
|
2.047
|
NM_032701
|
SUV420H2
|
suppressor of variegation 4-20 homolog 2 (Drosophila)
|
|
chr19_-_14108400
|
2.035
|
NM_018154
|
ASF1B
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae)
|
|
chr1_+_28458546
|
1.989
|
NM_031459
|
SESN2
|
sestrin 2
|
|
chr15_+_96304911
|
1.971
|
NM_183376
|
ARRDC4
|
arrestin domain containing 4
|
|
chr22_-_21193037
|
1.950
|
|
ZNF280B
|
zinc finger protein 280B
|
|
chr19_+_50373842
|
1.927
|
NM_212550
|
BLOC1S3
|
biogenesis of lysosomal organelles complex-1, subunit 3
|
|
chr5_+_151131655
|
1.922
|
NM_005754
NM_198395
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr17_+_54587641
|
1.920
|
NM_018304
|
PRR11
|
proline rich 11
|
|
chr22_+_20326591
|
1.918
|
|
SDF2L1
|
stromal cell-derived factor 2-like 1
|
|
chr16_-_2204776
|
1.916
|
NM_001042371
|
PGP
|
phosphoglycolate phosphatase
|
|
chr1_-_46370837
|
1.911
|
NM_003629
NM_001114172
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma)
|
|
chr20_-_60415729
|
1.905
|
NM_031215
|
CABLES2
|
Cdk5 and Abl enzyme substrate 2
|
|
chr2_+_69995807
|
1.886
|
|
MXD1
|
MAX dimerization protein 1
|
|
chr5_+_151131676
|
1.884
|
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr7_+_149707338
|
1.883
|
NM_173680
|
ZNF775
|
zinc finger protein 775
|
|
chr22_+_20326615
|
1.875
|
|
SDF2L1
|
stromal cell-derived factor 2-like 1
|
|
chr5_+_151131709
|
1.857
|
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr5_+_151131707
|
1.850
|
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr11_-_2677799
|
1.848
|
|
KCNQ1OT1
|
KCNQ1 overlapping transcript 1 (non-protein coding)
|
|
chr5_+_151131704
|
1.842
|
|
G3BP1
|
GTPase activating protein (SH3 domain) binding protein 1
|
|
chr17_-_54587306
|
1.824
|
|
SKA2
|
spindle and kinetochore associated complex subunit 2
|
|
chr1_-_151910098
|
1.811
|
NM_004515
|
ILF2
|
interleukin enhancer binding factor 2, 45kDa
|
|
chr17_+_43373887
|
1.793
|
NM_018129
|
PNPO
|
pyridoxamine 5'-phosphate oxidase
|
|
chr1_-_148936123
|
1.788
|
NM_018178
|
GOLPH3L
|
golgi phosphoprotein 3-like
|
|
chr6_-_31938530
|
1.768
|
|
NEU1
|
sialidase 1 (lysosomal sialidase)
|
|
chr1_+_210525490
|
1.753
|
NM_006243
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha
|
|
chr21_-_33774033
|
1.731
|
|
TMEM50B
|
transmembrane protein 50B
|
|
chr15_+_96304942
|
1.731
|
|
ARRDC4
|
arrestin domain containing 4
|
|
chr6_-_31890765
|
1.721
|
NM_005527
|
HSPA1L
|
heat shock 70kDa protein 1-like
|
|
chr22_+_36384647
|
1.681
|
NM_020315
|
PDXP
|
pyridoxal (pyridoxine, vitamin B6) phosphatase
|
|
chr21_-_33774096
|
1.680
|
|
TMEM50B
|
transmembrane protein 50B
|
|
chr7_-_72114342
|
1.673
|
NM_001013739
|
STAG3L3
STAG3L2
STAG3L1
|
stromal antigen 3-like 3
stromal antigen 3-like 2
stromal antigen 3-like 1
|
|
chr22_-_20551937
|
1.619
|
|
MAPK1
|
mitogen-activated protein kinase 1
|
|
chr6_+_31891293
|
1.605
|
|
HSPA1A
|
heat shock 70kDa protein 1A
|
|
chr21_-_33774150
|
1.596
|
NM_006134
|
TMEM50B
|
transmembrane protein 50B
|
|
chr6_+_31891509
|
1.595
|
|
HSPA1A
HSPA1B
|
heat shock 70kDa protein 1A
heat shock 70kDa protein 1B
|
|
chr20_+_61967033
|
1.595
|
NM_003288
NM_199359
NM_199360
NM_199361
NM_199362
NM_199363
|
TPD52L2
|
tumor protein D52-like 2
|
|
chr9_-_127043169
|
1.594
|
|
HSPA5
|
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa)
|
|
chr16_+_56327127
|
1.590
|
NM_005886
|
KATNB1
|
katanin p80 (WD repeat containing) subunit B 1
|
|
chr9_-_127043436
|
1.540
|
|
HSPA5
|
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa)
|
|
chr6_+_31891259
|
1.540
|
NM_005345
|
HSPA1A
|
heat shock 70kDa protein 1A
|
|
chr11_+_63710206
|
1.540
|
|
STIP1
|
stress-induced-phosphoprotein 1
|
|
chr22_-_20551953
|
1.536
|
NM_002745
NM_138957
|
MAPK1
|
mitogen-activated protein kinase 1
|
|
chr22_-_21193504
|
1.528
|
NM_080764
|
ZNF280B
|
zinc finger protein 280B
|
|
chr1_-_109742020
|
1.525
|
NM_002959
|
SORT1
|
sortilin 1
|
|
chr1_-_226679648
|
1.509
|
NM_003493
|
HIST3H3
|
histone cluster 3, H3
|
|
chr1_-_23567084
|
1.489
|
NM_030634
|
ZNF436
|
zinc finger protein 436
|
|
chr9_+_130883199
|
1.488
|
NM_001135917
NM_020438
|
DOLPP1
|
dolichyl pyrophosphate phosphatase 1
|
|
chr19_-_14108267
|
1.485
|
|
ASF1B
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae)
|
|
chr9_-_127043455
|
1.479
|
NM_005347
|
HSPA5
|
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa)
|
|
chr7_+_74826369
|
1.476
|
NM_001002840
NM_018991
|
STAG3L3
STAG3L2
STAG3L1
|
stromal antigen 3-like 3
stromal antigen 3-like 2
stromal antigen 3-like 1
|
|
chr1_-_159257486
|
1.476
|
|
F11R
|
F11 receptor
|
|
chr6_-_136613097
|
1.476
|
NM_001099286
NM_138419
|
FAM54A
|
family with sequence similarity 54, member A
|
|
chr2_-_73151894
|
1.466
|
|
SFXN5
|
sideroflexin 5
|
|
chr22_-_20551722
|
1.443
|
|
MAPK1
|
mitogen-activated protein kinase 1
|
|
chr7_+_100047660
|
1.425
|
NM_001040097
|
MOSPD3
|
motile sperm domain containing 3
|
|
chr7_-_100262971
|
1.406
|
NM_004444
|
EPHB4
|
EPH receptor B4
|
|
chr19_+_2928480
|
1.401
|
NM_001143986
NM_024760
|
TLE6
|
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila)
|
|
chr5_+_108112537
|
1.400
|
|
FER
|
fer (fps/fes related) tyrosine kinase
|
|
chr16_-_85908254
|
1.385
|
|
LOC100506581
|
hypothetical LOC100506581
|
|
chr11_+_63710242
|
1.358
|
|
STIP1
|
stress-induced-phosphoprotein 1
|
|
chr15_+_78139073
|
1.342
|
NM_019006
|
ZFAND6
|
zinc finger, AN1-type domain 6
|
|
chr19_+_18164989
|
1.323
|
NM_032683
|
MPV17L2
|
MPV17 mitochondrial membrane protein-like 2
|
|
chr1_+_110683580
|
1.316
|
|
RBM15
|
RNA binding motif protein 15
|
|
chr15_+_78139098
|
1.301
|
|
ZFAND6
|
zinc finger, AN1-type domain 6
|
|
chr9_+_132990760
|
1.290
|
NM_005085
|
NUP214
|
nucleoporin 214kDa
|
|
chr20_+_61967078
|
1.269
|
|
TPD52L2
|
tumor protein D52-like 2
|
|
chr9_+_132990834
|
1.265
|
|
NUP214
|
nucleoporin 214kDa
|
|
chr1_+_148726497
|
1.264
|
|
TARS2
|
threonyl-tRNA synthetase 2, mitochondrial (putative)
|
|
chr1_+_150029798
|
1.259
|
|
|
|
|
chr17_+_1566566
|
1.253
|
NM_001163673
NM_152348
|
WDR81
|
WD repeat domain 81
|
|
chr4_-_2233504
|
1.249
|
NM_006454
|
MXD4
|
MAX dimerization protein 4
|
|
chr16_-_65774904
|
1.236
|
|
KIAA0895L
|
KIAA0895-like
|
|
chr6_-_33493640
|
1.232
|
|
CUTA
|
cutA divalent cation tolerance homolog (E. coli)
|
|
chr1_+_6767968
|
1.231
|
NM_001195563
NM_015215
|
CAMTA1
|
calmodulin binding transcription activator 1
|
|
chr19_+_19035792
|
1.223
|
NM_178526
|
SLC25A42
|
solute carrier family 25, member 42
|
|
chr16_+_2419388
|
1.208
|
NM_001761
|
CCNF
|
cyclin F
|
|
chr14_-_22825148
|
1.197
|
NM_020834
|
HOMEZ
|
homeobox and leucine zipper encoding
|
|
chr17_+_71772875
|
1.190
|
NM_182565
|
FAM100B
|
family with sequence similarity 100, member B
|
|
chr1_-_180627512
|
1.190
|
|
GLUL
|
glutamate-ammonia ligase
|
|
chr1_+_110683586
|
1.183
|
|
RBM15
|
RNA binding motif protein 15
|
|
chr5_+_44844724
|
1.182
|
NM_016640
|
MRPS30
|
mitochondrial ribosomal protein S30
|
|
chr5_+_122875691
|
1.178
|
NM_001031812
NM_001044722
NM_001044723
NM_004384
|
CSNK1G3
|
casein kinase 1, gamma 3
|
|
chr2_-_73152323
|
1.177
|
NM_144579
|
SFXN5
|
sideroflexin 5
|
|
chr10_+_62208094
|
1.173
|
NM_001170406
NM_001170407
NM_001786
NM_033379
|
CDK1
|
cyclin-dependent kinase 1
|
|
chr10_+_70418482
|
1.165
|
NM_015634
|
KIAA1279
|
KIAA1279
|
|
chr1_-_28113540
|
1.162
|
|
RPA2
|
replication protein A2, 32kDa
|
|
chr17_-_77411803
|
1.162
|
NM_000918
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr2_-_136005238
|
1.161
|
NM_032143
|
ZRANB3
|
zinc finger, RAN-binding domain containing 3
|
|
chr9_-_139604720
|
1.153
|
NM_138462
|
ZMYND19
|
zinc finger, MYND-type containing 19
|
|
chr19_+_38154978
|
1.153
|
NM_152266
|
C19orf40
|
chromosome 19 open reading frame 40
|
|
chr6_+_16237264
|
1.146
|
NM_013262
|
MYLIP
|
myosin regulatory light chain interacting protein
|
|
chr1_-_150029529
|
1.144
|
NM_001083963
NM_001083964
NM_001083965
NM_006862
|
TDRKH
|
tudor and KH domain containing
|
|
chr16_+_2419418
|
1.131
|
|
CCNF
|
cyclin F
|
|
chr7_+_98940801
|
1.126
|
|
ZKSCAN5
|
zinc finger with KRAB and SCAN domains 5
|
|
chr1_-_195382189
|
1.124
|
NM_018136
|
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila)
|
|
chr14_-_22825072
|
1.124
|
|
HOMEZ
|
homeobox and leucine zipper encoding
|
|
chr19_+_63482118
|
1.123
|
NM_021089
|
ZNF8
|
zinc finger protein 8
|
|
chr11_+_63710162
|
1.123
|
NM_006819
|
STIP1
|
stress-induced-phosphoprotein 1
|
|
chr6_+_16237300
|
1.123
|
|
MYLIP
|
myosin regulatory light chain interacting protein
|
|
chr17_+_4560680
|
1.119
|
|
ARRB2
|
arrestin, beta 2
|
|
chr1_-_159257595
|
1.113
|
NM_016946
|
F11R
|
F11 receptor
|
|
chr17_+_4560529
|
1.112
|
NM_004313
NM_199004
|
ARRB2
|
arrestin, beta 2
|
|
chr4_-_2233606
|
1.110
|
|
MXD4
|
MAX dimerization protein 4
|
|
chr4_-_1683712
|
1.110
|
NM_006527
|
SLBP
|
stem-loop binding protein
|
|
chr14_-_72430545
|
1.108
|
NM_012074
|
DPF3
|
D4, zinc and double PHD fingers, family 3
|
|
chr17_+_71772982
|
1.102
|
|
FAM100B
|
family with sequence similarity 100, member B
|
|
chr22_-_17546296
|
1.092
|
NM_005984
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1
|
|
chr7_-_66404827
|
1.089
|
|
PMS2P4
|
postmeiotic segregation increased 2 pseudogene 4
|
|
chr7_-_73944616
|
1.089
|
|
STAG3L2
|
stromal antigen 3-like 2
|
|
chr20_+_61967094
|
1.088
|
|
TPD52L2
|
tumor protein D52-like 2
|
|
chr15_+_78139018
|
1.078
|
|
ZFAND6
|
zinc finger, AN1-type domain 6
|
|
chr16_-_85908448
|
1.076
|
NM_001195124
NM_001195125
|
LOC100506581
|
hypothetical LOC100506581
|
|
chr7_-_137337318
|
1.075
|
|
CREB3L2
|
cAMP responsive element binding protein 3-like 2
|
|
chr17_-_77411733
|
1.071
|
|
P4HB
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr7_+_129497572
|
1.069
|
NM_014997
|
KLHDC10
|
kelch domain containing 10
|
|
chr1_+_5975286
|
1.066
|
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr7_-_137337352
|
1.065
|
|
CREB3L2
|
cAMP responsive element binding protein 3-like 2
|
|
chr1_-_159257567
|
1.063
|
|
F11R
|
F11 receptor
|
|
chr22_-_17546276
|
1.061
|
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1
|
|
chr1_+_144149794
|
1.056
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr7_-_150305933
|
1.044
|
NM_000238
NM_172056
|
KCNH2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2
|
|
chr7_+_100047877
|
1.034
|
NM_001040098
NM_001040099
NM_023948
|
MOSPD3
|
motile sperm domain containing 3
|
|
chr4_+_38860418
|
1.031
|
NM_025132
|
WDR19
|
WD repeat domain 19
|
|
chr1_+_5975345
|
1.024
|
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr6_-_32253819
|
1.024
|
NM_032741
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha)
|
|
chr14_+_53933422
|
1.023
|
NM_001130851
NM_005192
|
CDKN3
|
cyclin-dependent kinase inhibitor 3
|
|
chr12_-_120725210
|
1.022
|
|
LOC338799
|
hypothetical LOC338799
|
|
chr7_+_66405077
|
1.020
|
|
STAG3L4
|
stromal antigen 3-like 4
|
|
chrX_+_48934831
|
1.019
|
|
|
|
|
chr17_+_56032385
|
1.014
|
|
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D
|
|
chr7_+_66405010
|
1.008
|
NM_022906
|
STAG3L4
|
stromal antigen 3-like 4
|
|
chr7_-_129868059
|
1.002
|
NM_018718
|
TSGA14
|
testis specific, 14
|
|
chr7_+_66405122
|
0.999
|
|
STAG3L4
|
stromal antigen 3-like 4
|
|
chr22_-_17546234
|
0.994
|
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1
|
|
chr7_+_73944831
|
0.990
|
|
PMS2P5
|
postmeiotic segregation increased 2 pseudogene 5
|
|
chr1_-_148124738
|
0.988
|
NM_003528
|
HIST2H2BE
|
histone cluster 2, H2be
|
|
chr21_+_42946719
|
0.979
|
NM_001001567
NM_001001568
NM_001001569
NM_001001570
NM_001001571
NM_001001572
NM_001001573
NM_001001574
NM_001001575
NM_001001576
NM_001001577
NM_001001578
NM_001001579
NM_001001580
NM_001001581
NM_001001582
NM_001001583
NM_001001584
NM_001001585
NM_002606
|
PDE9A
|
phosphodiesterase 9A
|
|
chr1_-_52116598
|
0.977
|
|
NRD1
|
nardilysin (N-arginine dibasic convertase)
|
|
chr17_-_70797051
|
0.976
|
|
SLC25A19
|
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19
|
|
chr1_-_11245193
|
0.960
|
NM_004958
|
MTOR
|
mechanistic target of rapamycin (serine/threonine kinase)
|
|
chr15_+_86965526
|
0.959
|
NM_022767
|
AEN
|
apoptosis enhancing nuclease
|
|
chr1_-_11245181
|
0.956
|
|
MTOR
|
mechanistic target of rapamycin (serine/threonine kinase)
|
|
chr9_-_37455429
|
0.955
|
|
ZBTB5
|
zinc finger and BTB domain containing 5
|
|
chr19_-_53832381
|
0.951
|
NM_001352
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein
|
|
chr19_-_38154631
|
0.950
|
NM_032816
|
CCDC123
|
coiled-coil domain containing 123
|
|
chr17_+_43374021
|
0.950
|
|
PNPO
|
pyridoxamine 5'-phosphate oxidase
|
|
chr1_-_11245130
|
0.949
|
|
MTOR
|
mechanistic target of rapamycin (serine/threonine kinase)
|
|
chr14_-_103251497
|
0.947
|
NM_001100118
NM_001100119
NM_005432
|
XRCC3
|
X-ray repair complementing defective repair in Chinese hamster cells 3
|
|
chr1_+_148726537
|
0.946
|
NM_025150
|
TARS2
|
threonyl-tRNA synthetase 2, mitochondrial (putative)
|
|
chr1_-_171838802
|
0.941
|
NM_178527
|
SLC9A11
|
solute carrier family 9, member 11
|
|
chr1_+_170017430
|
0.940
|
|
METTL13
|
methyltransferase like 13
|
|
chr22_+_20316968
|
0.939
|
NM_152612
|
CCDC116
|
coiled-coil domain containing 116
|
|
chrX_+_48265107
|
0.937
|
NM_006579
|
EBP
|
emopamil binding protein (sterol isomerase)
|
|
chr1_+_170017452
|
0.937
|
|
METTL13
|
methyltransferase like 13
|
|
chr17_+_58915909
|
0.936
|
NM_001178057
NM_152830
|
ACE
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1
|
|
chr1_-_19101603
|
0.933
|
NM_003748
NM_170726
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1
|
|
chr1_-_11245172
|
0.933
|
|
MTOR
|
mechanistic target of rapamycin (serine/threonine kinase)
|
|
chr7_+_98940537
|
0.930
|
|
ZKSCAN5
|
zinc finger with KRAB and SCAN domains 5
|
|
chr19_+_50887580
|
0.928
|
NM_001163377
NM_017659
|
QPCTL
|
glutaminyl-peptide cyclotransferase-like
|
|
chr19_-_6375536
|
0.925
|
|
KHSRP
|
KH-type splicing regulatory protein
|
|
chr6_-_32203971
|
0.921
|
NM_001136153
NM_004381
|
ATF6B
|
activating transcription factor 6 beta
|
|
chr19_+_41298493
|
0.919
|
|
TBCB
|
tubulin folding cofactor B
|
|
chrX_-_48844929
|
0.919
|
NM_007075
|
WDR45
|
WD repeat domain 45
|
|
chr17_+_38576707
|
0.918
|
NM_005899
|
NBR1
|
neighbor of BRCA1 gene 1
|
|
chr7_-_100021658
|
0.917
|
|
LRCH4
|
leucine-rich repeats and calponin homology (CH) domain containing 4
|
|
chr10_-_69267774
|
0.913
|
NM_021800
NM_201262
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12
|
|
chr1_+_24158887
|
0.913
|
NM_017761
|
PNRC2
|
proline-rich nuclear receptor coactivator 2
|
|
chr11_-_64302440
|
0.913
|
|
SF1
|
splicing factor 1
|
|
chr16_-_45280612
|
0.912
|
NM_018206
|
VPS35
|
vacuolar protein sorting 35 homolog (S. cerevisiae)
|
|
chr19_-_10352212
|
0.908
|
NM_003331
|
TYK2
|
tyrosine kinase 2
|
|
chr1_+_42920889
|
0.905
|
|
YBX1
|
Y box binding protein 1
|
|
chr1_-_112083387
|
0.897
|
NM_019099
|
C1orf183
|
chromosome 1 open reading frame 183
|
|
chr3_-_37009571
|
0.893
|
|
|
|
|
chr1_-_28113817
|
0.892
|
NM_002946
|
RPA2
|
replication protein A2, 32kDa
|
|
chr1_+_24158898
|
0.891
|
|
PNRC2
|
proline-rich nuclear receptor coactivator 2
|